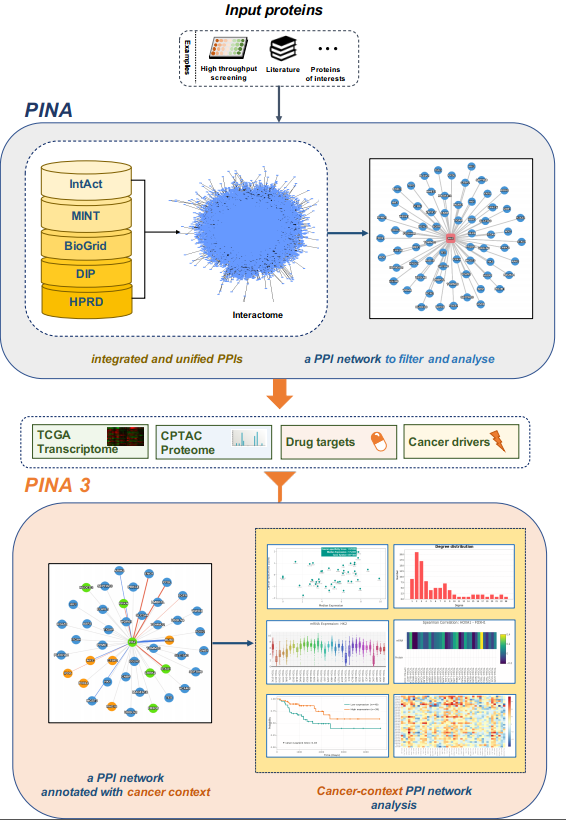
Protein Interaction Network Analysis (PINA) platform is an integrated platform for protein interaction network construction, filtering, analysis, visualization and management. It integrates protein-protein interaction data from six public curated databases and builds a complete, non-redundant protein interaction dataset for six model organisms. Moreover, it provides a variety of built-in tools to filter, visualize and analyze interaction networks for gaining biological insights at network level.
PINA 3.0 was released in 2020 to help revealing tumour type-specific insights from PPI networks. It provides a number of new analysis and visualization utilities by integrating PPI data with TCGA and CPTAC datasets, to help characterize the cancer context for a PPI network, including:
- to infer network proteins with tumor type expression specificity.
- to identify candidate prognosis biomarkers, putative mutational cancer drivers, and therapeutic targets in a network, for a specific cancer type.
- to identify pairs of co-expressing interacting proteins across cancer types.
Further reading: PINA Tutorial and recent publication at Nucleic Acids Research [Full text].
Start your trip with PINA 3.0 from here.