Introduction
Reproducible Prognosis Molecular Signature (ReProMSig) platform could help develop and validate a multivariable prognostic/predictive biomarker in a transparent and reproducible way, with the following advanced features:
- It streamlines the analysis process in development of a multivariable prediction model using molecular profiles and/or clinicopathological factors, as well as evaluation of its prognostic and/or predictive value.
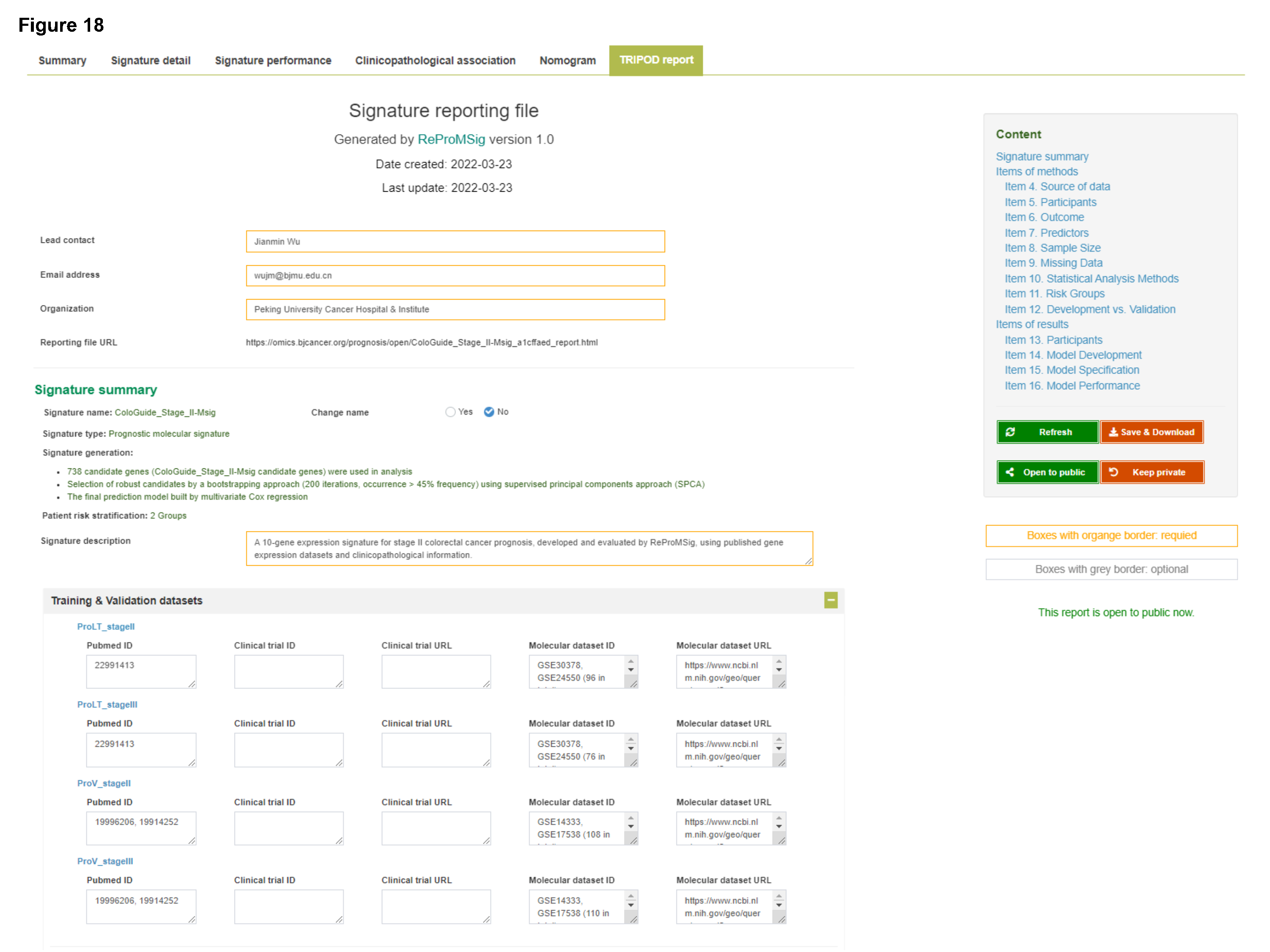
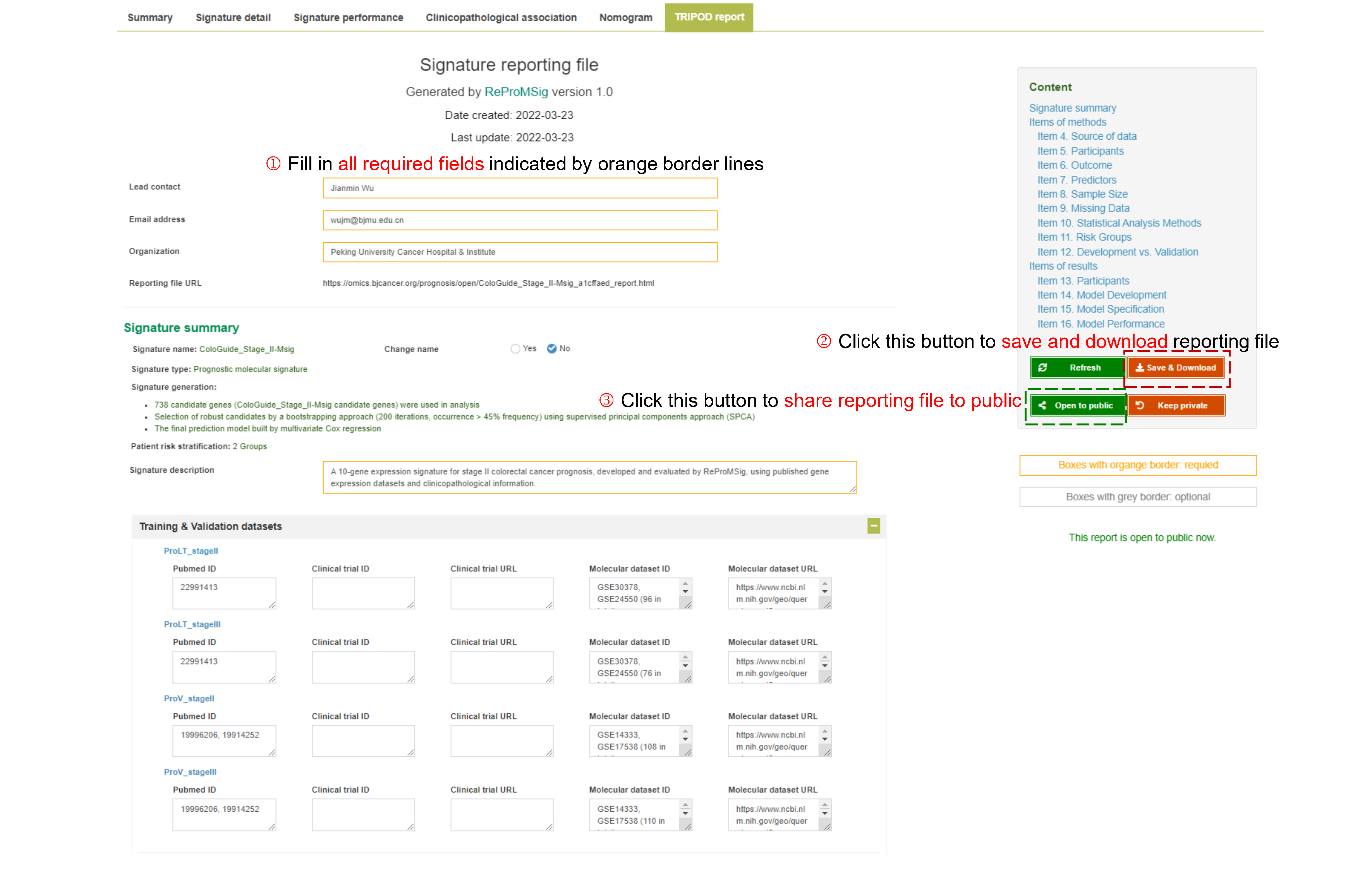
- The full detail of modelling procedures and results can be provided as a signature report file (example report), which is well-designed following the TRIPOD statement.
- Long-term storage and management of user datasets and signatures are supported for registered users.
- Risk assessment for single patient using a developed biomarker is provided for research purpose.
- A standalone version is available at GitHub for experienced users to perform local analysis.

This module allows users to upload unpublished datasets for signature development and validation. Uploaded datasets will be searchable in the Signature Development module, to the uploader user only.

This module allows management of developed private signatures and generation of signature report file, which includes comprehensive methodology details and visual results, in a structure following TRIPOD guideline check items. In addition, public signatures and associated signature reports are supported for sharing with the community.
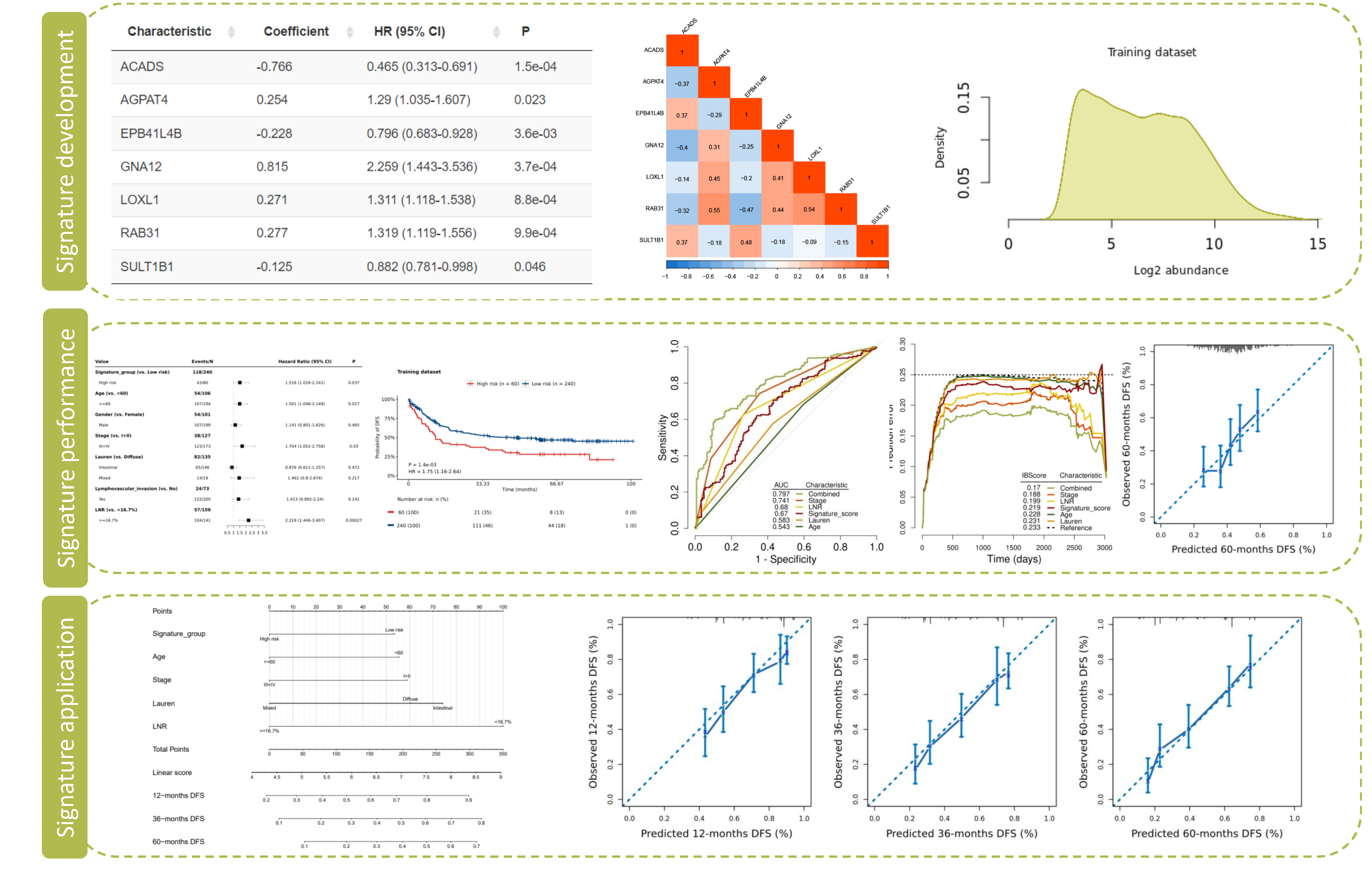
This module implements the complete analysis process in a streamlined way utilizing private/published datasets, to develop a multivariable prognosis prediction model with or without molecular signature, to validate the prognosis model in internal and external datasets, and to assess the performance thoroughly.

This module calculates the risk score and event probabilities at specified intervals for a new case, using a private/public prognostic/predictive signature, and assesses patient risk or treatment benefit correspondingly.
Registering a user account will support long-term storage of developed signatures and uploaded datasets. It also allows registered users to share the generated model reporting files with the community for transparency and reproducibility.

Before using this platform, please read this statement carefully and thoroughly. By using this platform, you are indicating your assent to the entire content of this statement.
Copyright © 2023
Center for Cancer Bioinformatics
All Rights Reserved.
Peking University Cancer Hospital & Institute, Peking University Health Science Center

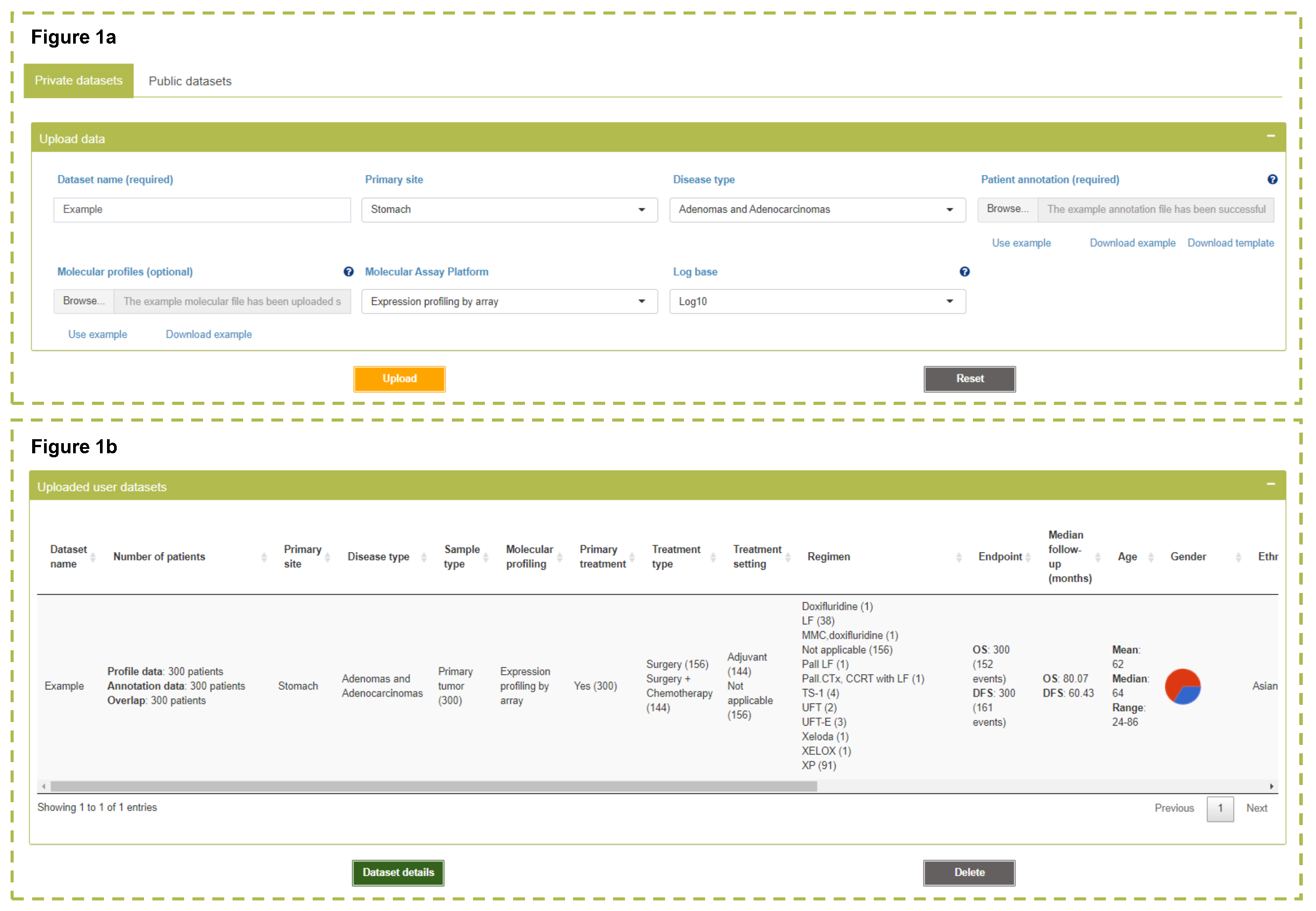
My datasets
ReProMSig allows users to upload unpublished datasets for development and validation of prognosis prediction models. The uploaded datasets can be used for Signature Development or Single Patient Prediction analysis.
Please note:
- A full document of file format is described in the tutorial.
- Registered users can save their datasets in the platform for future access and analysis. The uploaded datasets by unregistered users will be removed from the platform after disconnection, or manually refreshing the web page.
Copyright © 2023
Center for Cancer Bioinformatics
All Rights Reserved.
Peking University Cancer Hospital & Institute, Peking University Health Science Center

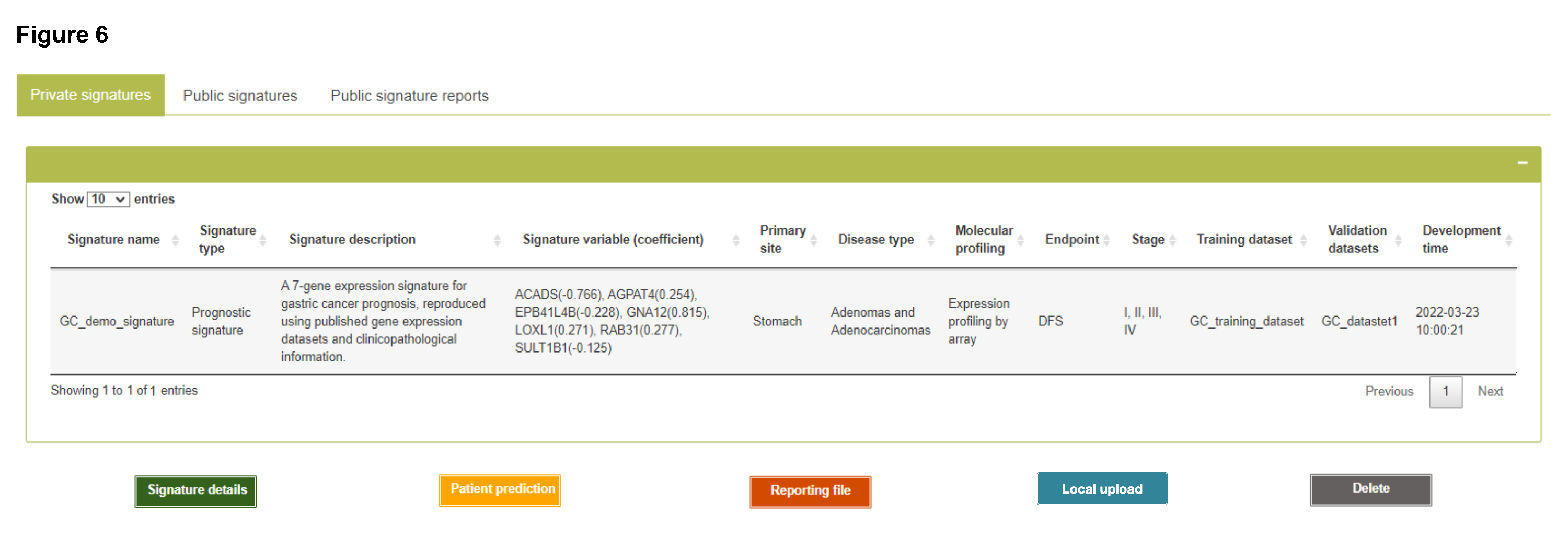
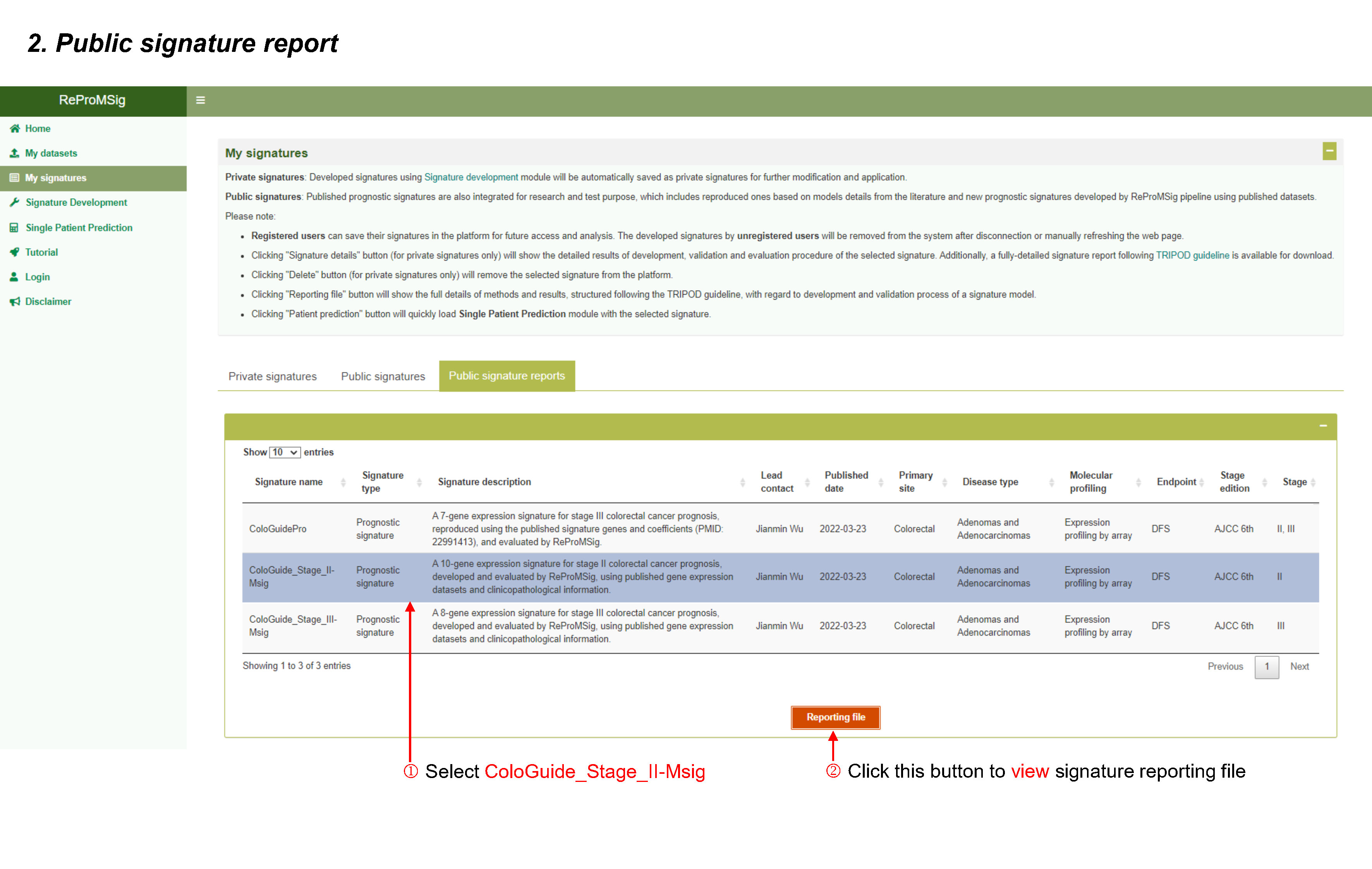
My signatures
Private signatures: Developed signatures using Signature development module will be automatically saved as private signatures for further modification and application.
Public signatures: Published prognostic signatures are also integrated for research and test purpose, which includes reproduced ones based on models details from the literature and new prognostic signatures developed by ReProMSig pipeline using published datasets.
Please note:
- Registered users can save their signatures in the platform for future access and analysis. The developed signatures by unregistered users will be removed from the system after disconnection or manually refreshing the web page.
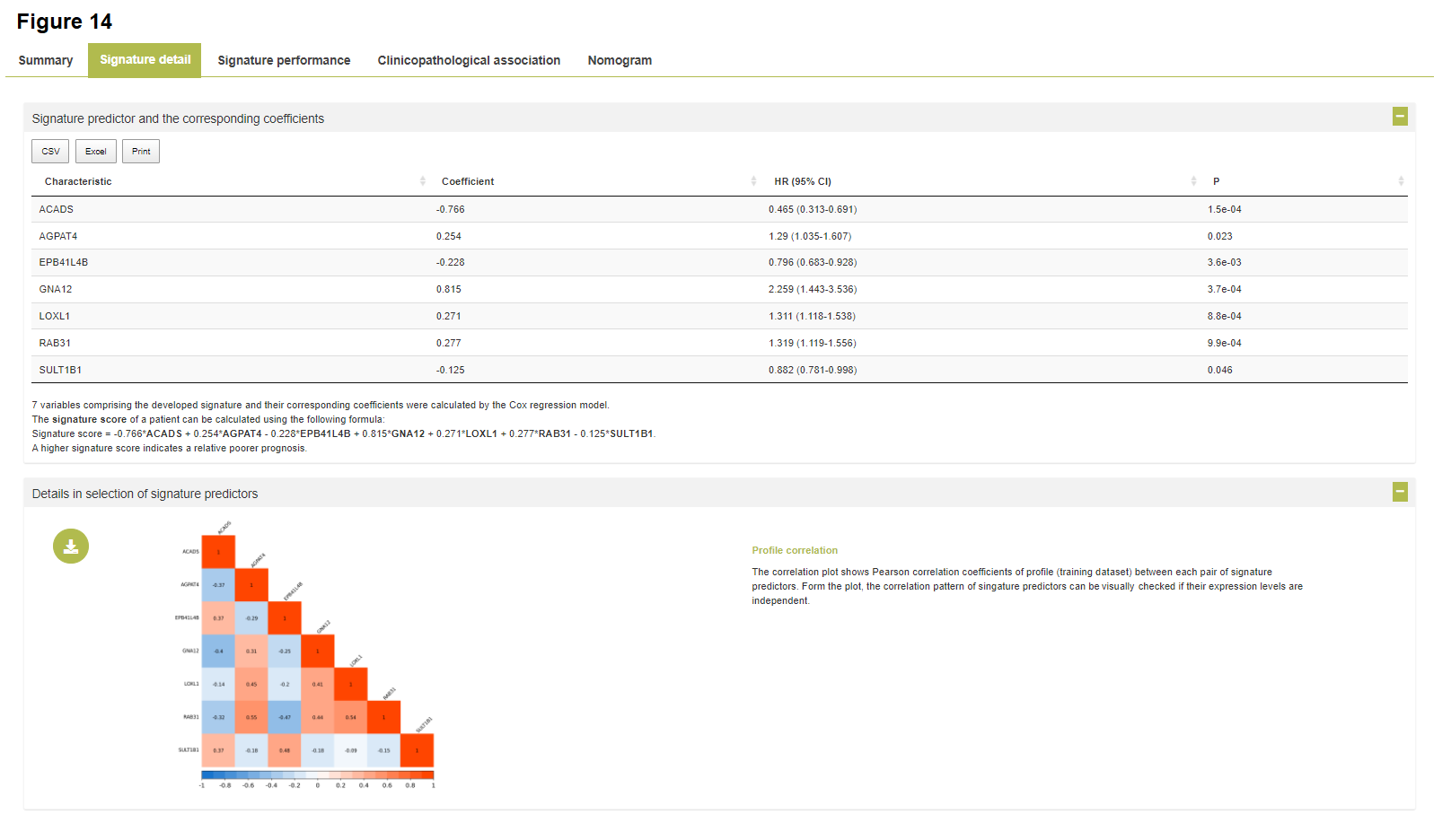
- Clicking "Signature details" button (for private signatures only) will show the detailed results of development, validation and evaluation procedure of the selected signature. Additionally, a fully-detailed signature report following TRIPOD guideline is available for download.
- Clicking "Delete" button (for private signatures only) will remove the selected signature from the platform.
- Clicking "Reporting file" button will show the full details of methods and results, structured following the TRIPOD guideline, with regard to development and validation process of a signature model.
- Clicking "Patient prediction" button will quickly load Single Patient Prediction module with the selected signature.
- Clicking "Local upload" button allows users to upload local signature model and reporting file that were generated using the standalone version of ReProMSig (https://github.com/WuOmicsLab/ReProMSig).
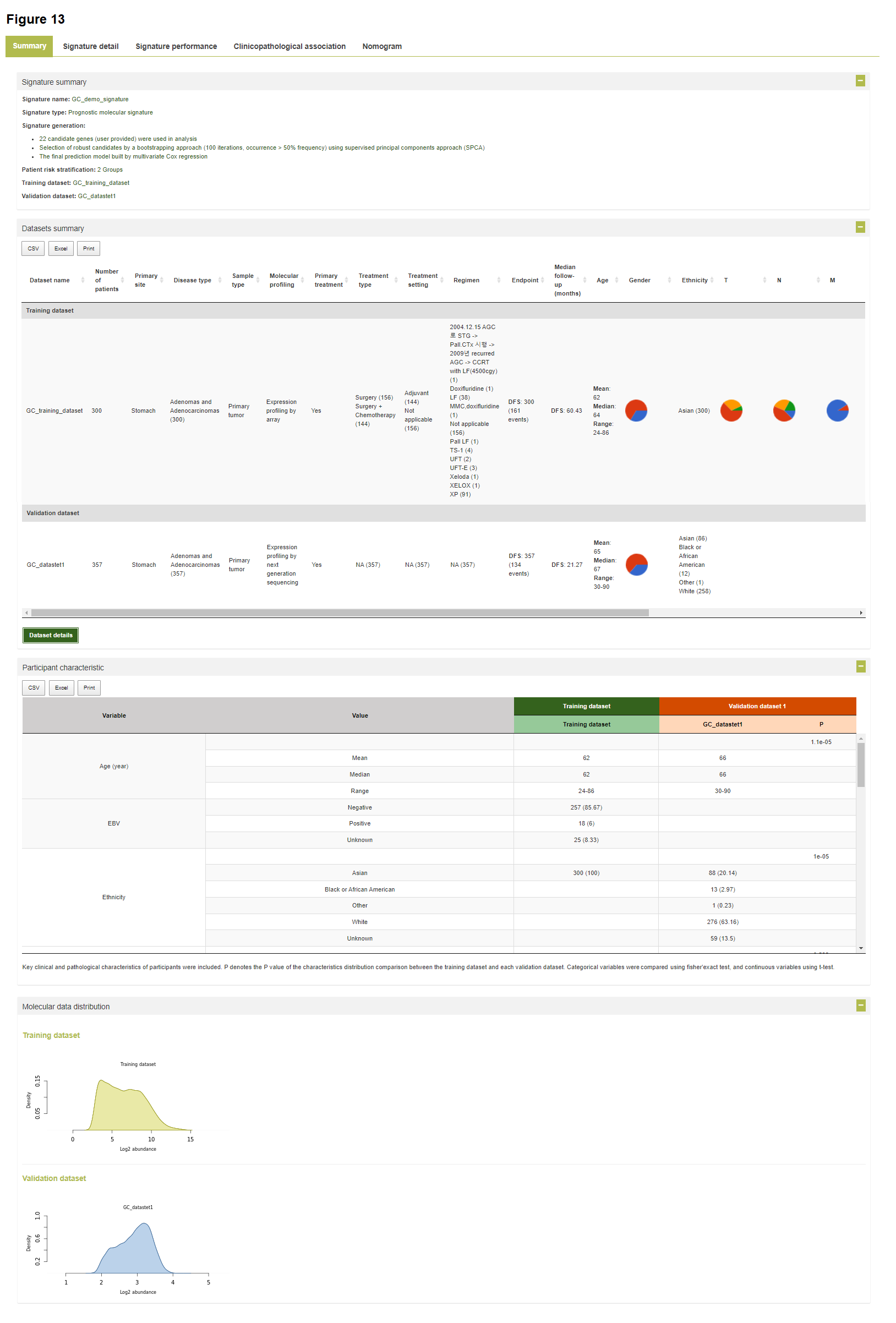
Results
The results of signature development, validation and evaluation are categorized in multiple tabs as below:
My signatures
Private signatures: Developed signatures using Signature development module will be automatically saved as private signatures for further modification and application.
Public signatures: Published prognostic signatures are also integrated for research and test purpose, which includes reproduced ones based on models details from the literature and new prognostic signatures developed by ReProMSig pipeline using published datasets.
Please note:
- Registered users can save their signatures in the platform for future access and analysis. The developed signatures by unregistered users will be removed from the system after disconnection or manually refreshing the web page.
- Clicking "Signature details" button (for private signatures only) will show the detailed results of development, validation and evaluation procedure of the selected signature. Additionally, a fully-detailed signature report following TRIPOD guideline is available for download.
- Clicking "Delete" button (for private signatures only) will remove the selected signature from the platform.
- Clicking "Reporting file" button will show the full details of methods and results, structured following the TRIPOD guideline, with regard to development and validation process of a signature model.
- Clicking "Patient prediction" button will quickly load Single Patient Prediction module with the selected signature.
- Clicking "Local upload" button allows users to upload local signature model and reporting file that were generated using the standalone version of ReProMSig (https://github.com/WuOmicsLab/ReProMSig).
Upload local signature
Rdata file
Reporting file
Copyright © 2023
Center for Cancer Bioinformatics
All Rights Reserved.
Peking University Cancer Hospital & Institute, Peking University Health Science Center

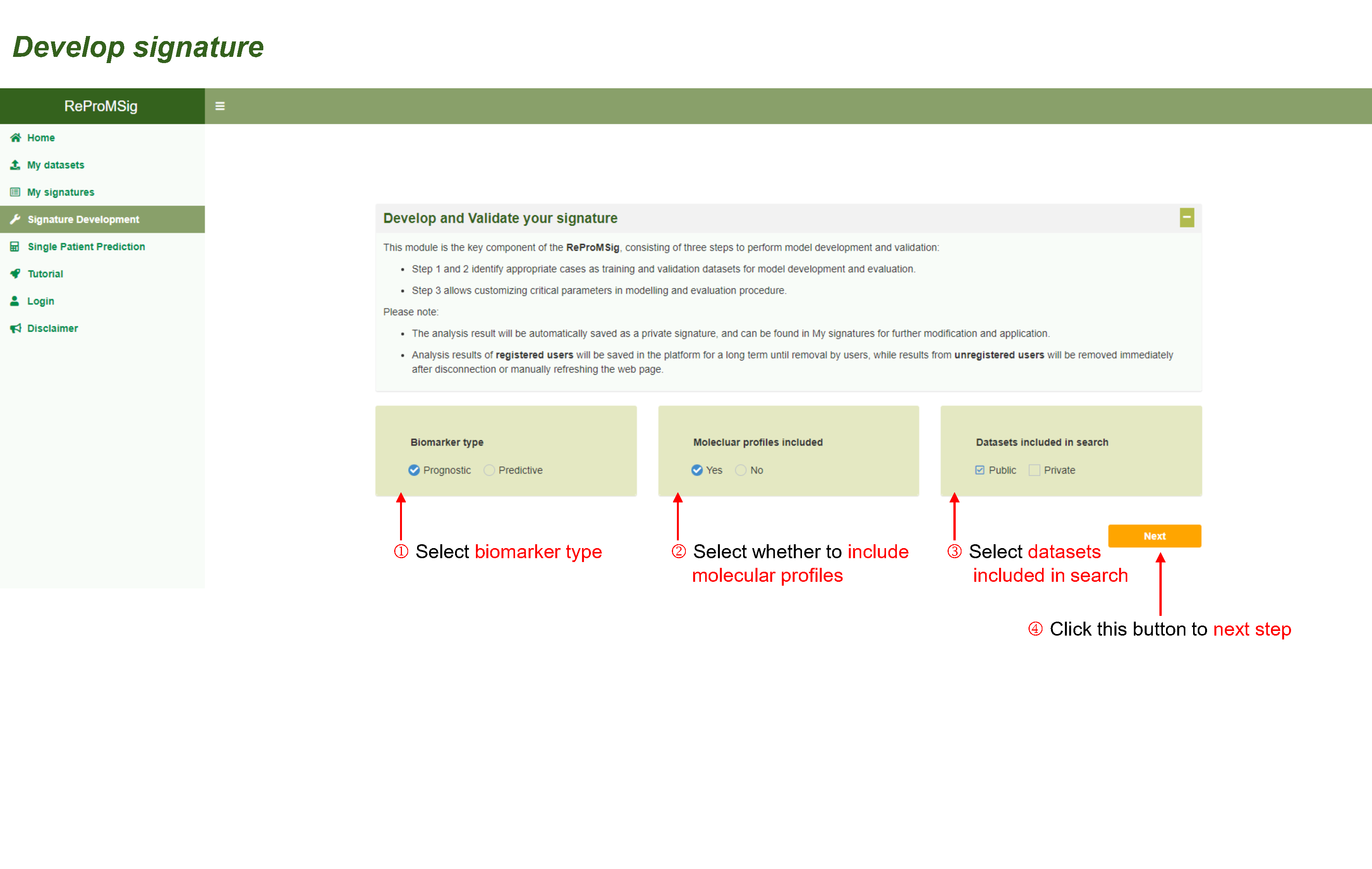
Develop and Validate your signature
This module is the key component of the ReProMSig, consisting of three steps to perform model development and validation:
- Step 1 and 2 identify appropriate cases as training and validation datasets for model development and evaluation.
- Step 3 allows customizing critical parameters in modelling and evaluation procedure.
Please note:
- The analysis result will be automatically saved as a private signature, and can be found in My signatures for further modification and application.
- Analysis results of registered users will be saved in the platform for a long term until removal by users, while results from unregistered users will be removed immediately after disconnection or manually refreshing the web page.
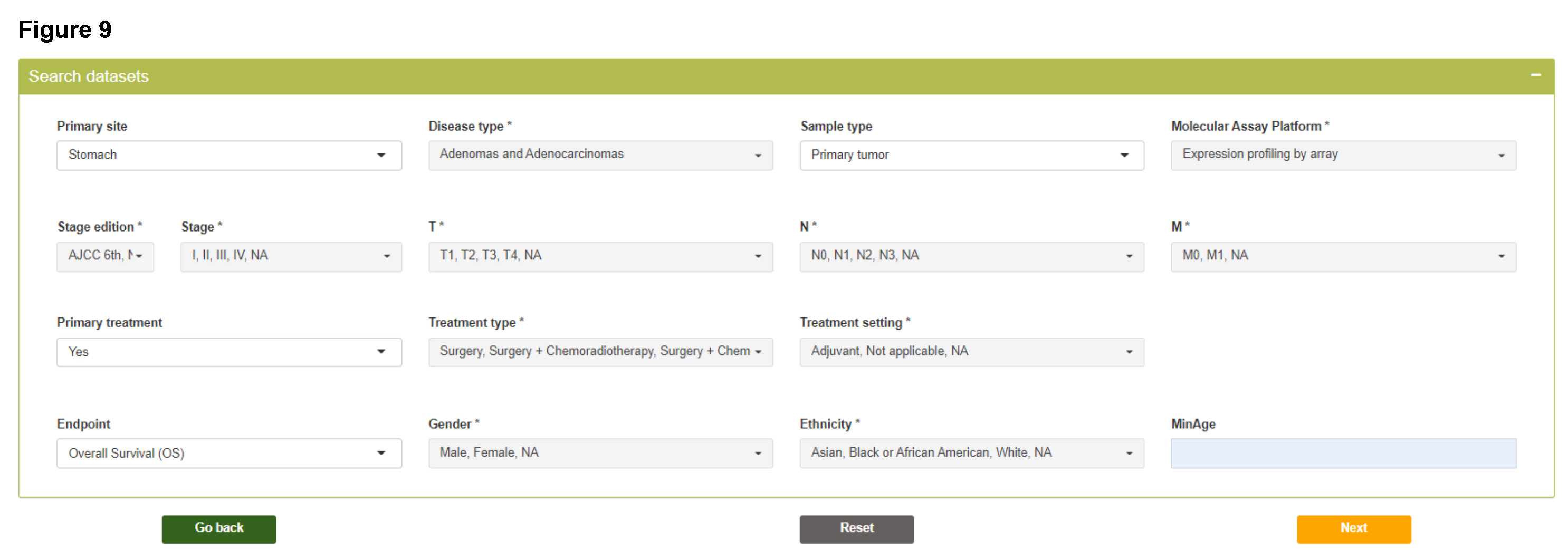
Step 1
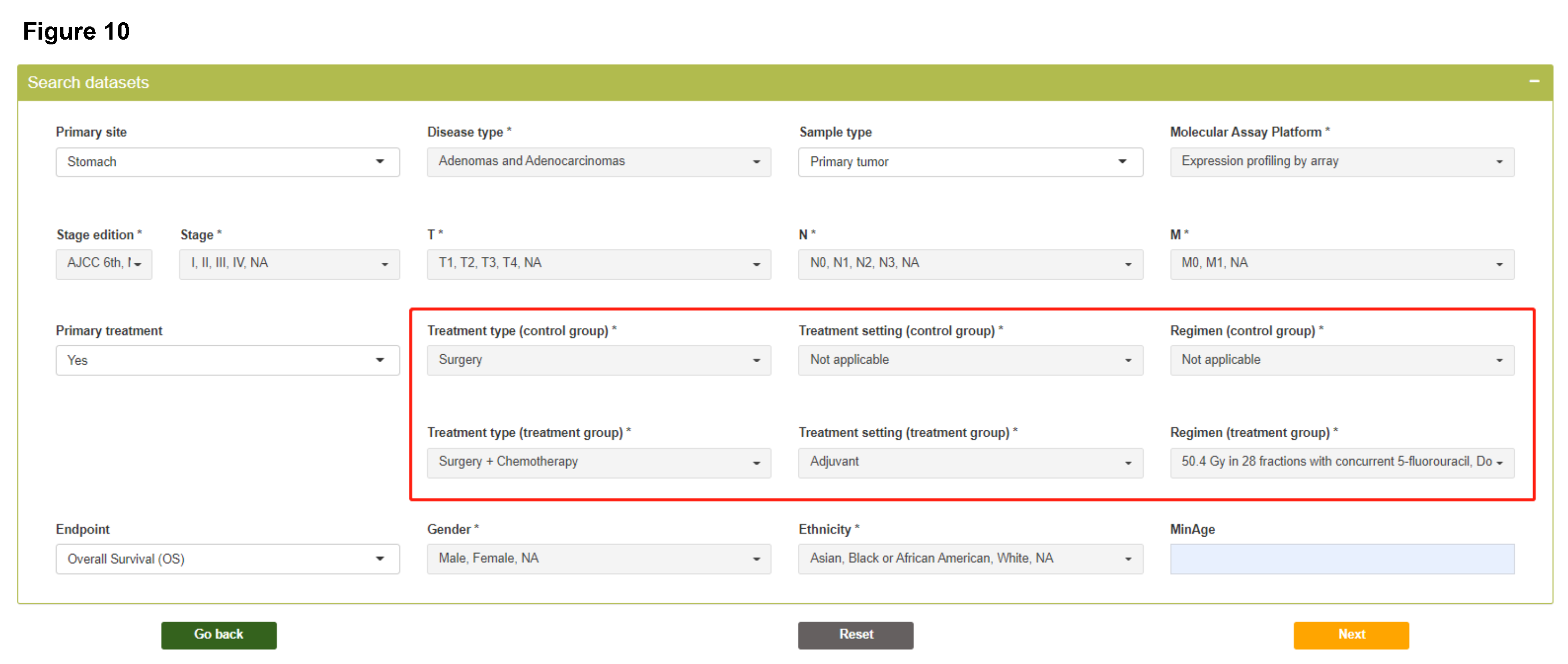
As the first step, histological and clinicopathological variables need to be specified according to research questions, which will be used to identify patients meeting query criteria from different datasets.
- Please note that variables with * sign allow multiple selection in the dropdown list.
- The options of "Primary site", "Disease type", "Sample type", "Treatment type", "Treatment setting", "Regimen", and "Gender" are automatically obtained from the accessible datasets included in the last step.
Search datasets
Step 2
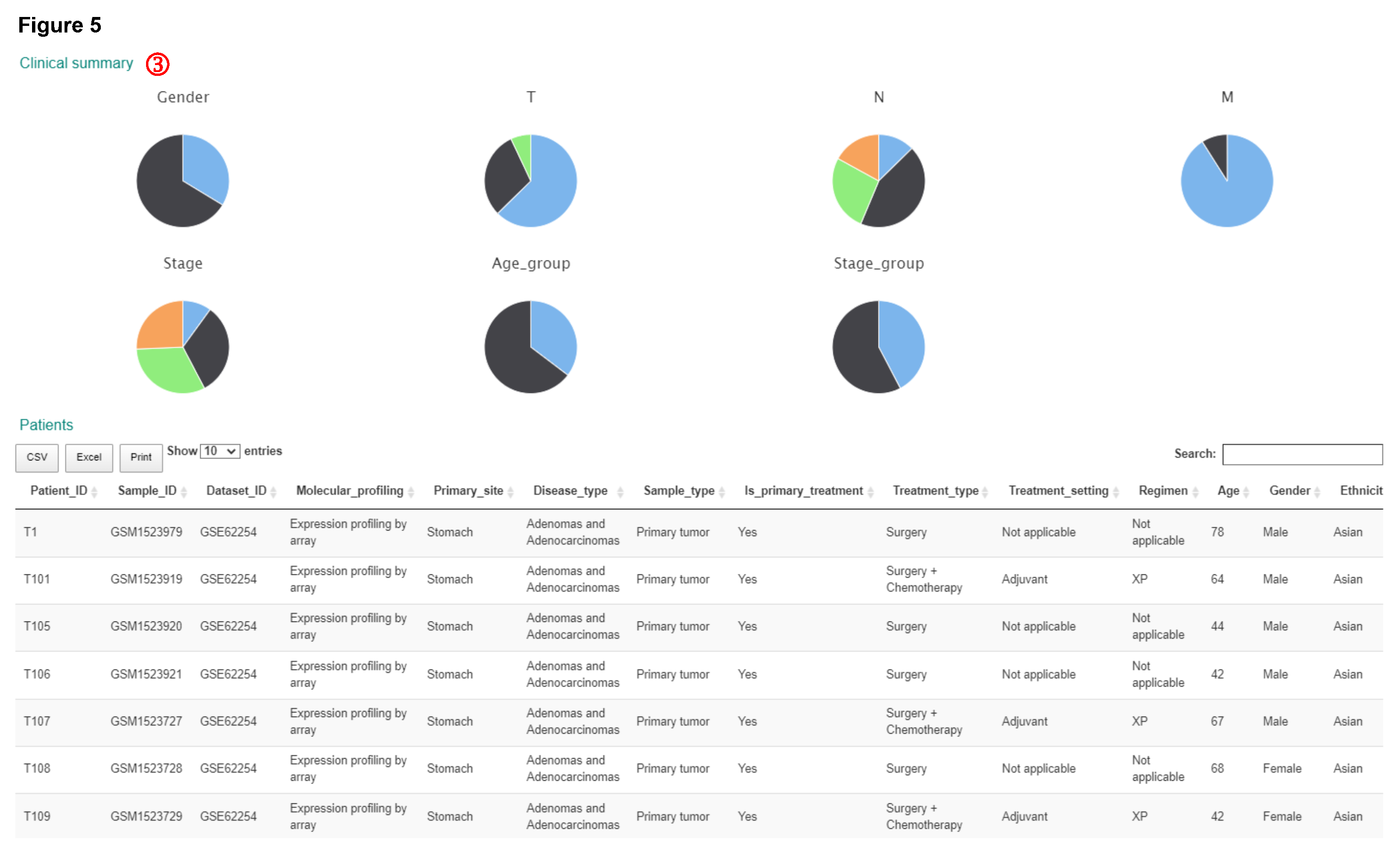
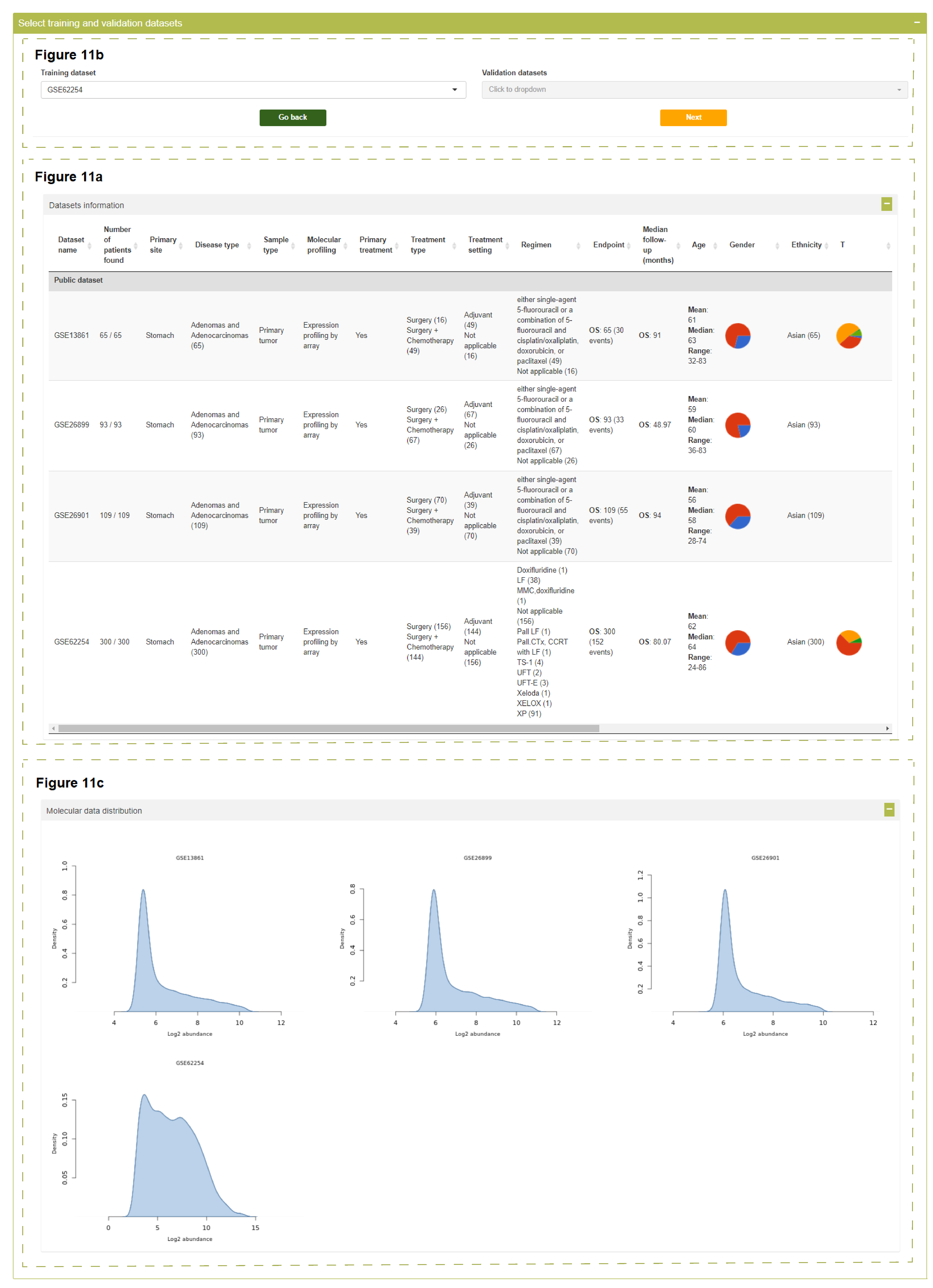
Statistics of samples meeting the query criteria are summarized for each dataset and listed in the interactive table shown below.
Please note:
- In the "Number of patients found" column, the number of patients meeting the query criteria and the number of all patients in this dataset are separated by a slash.
- You need to choose a dataset used for model training. Optionally, select one or more datasets used for external validation, which dropdown list allows multiple selection.
Select training and validation datasets
Datasets information
Molecular data distribution
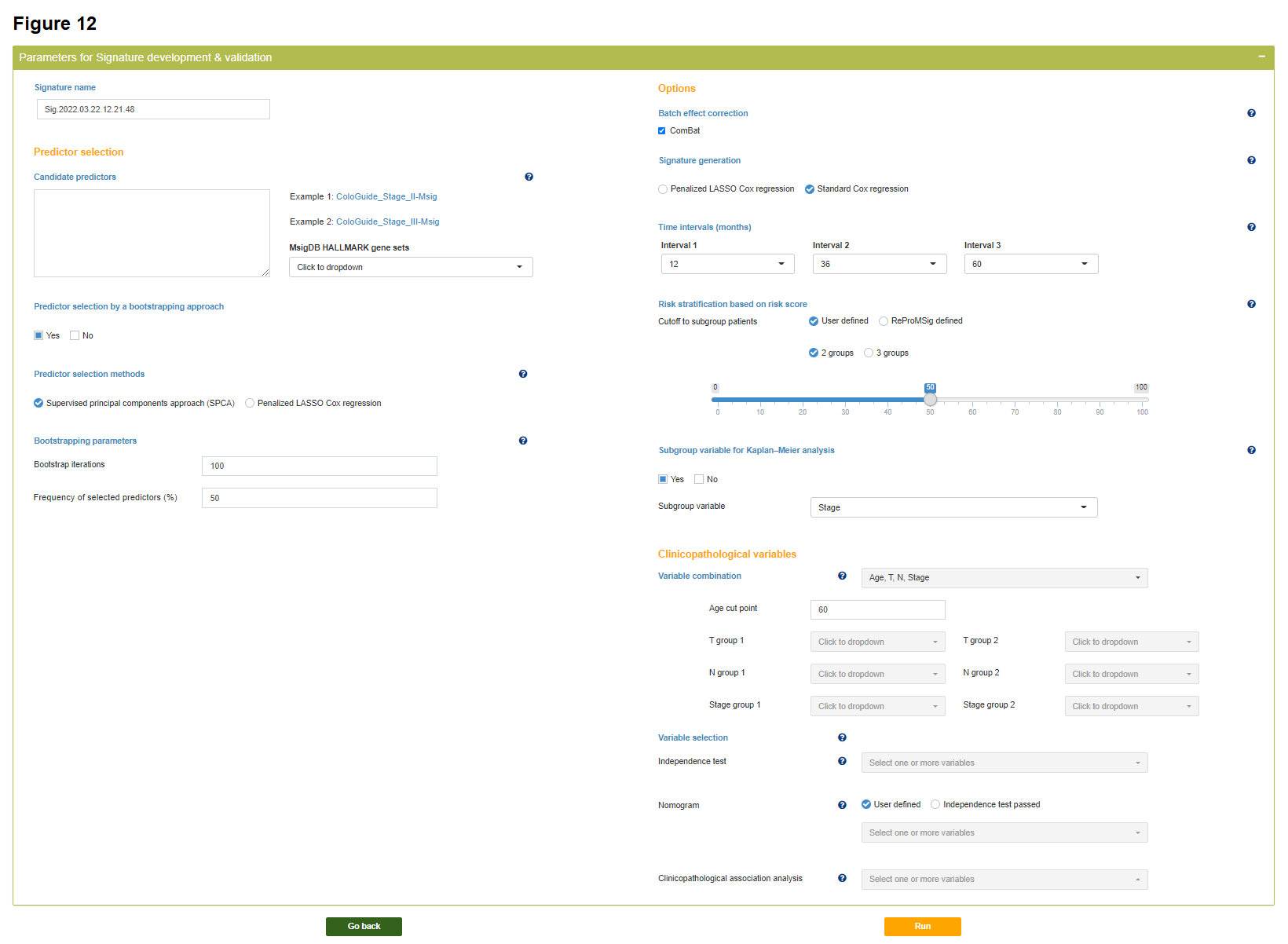
Step 3
Users can specify the key parameters in the procedure of development, validation and evaluation of a prognosis model. Details of each parameter will be shown by clicking the nearby question mark icon.
Parameters for signature development & validation
Signature name
Predictor selection
Candidate predictors
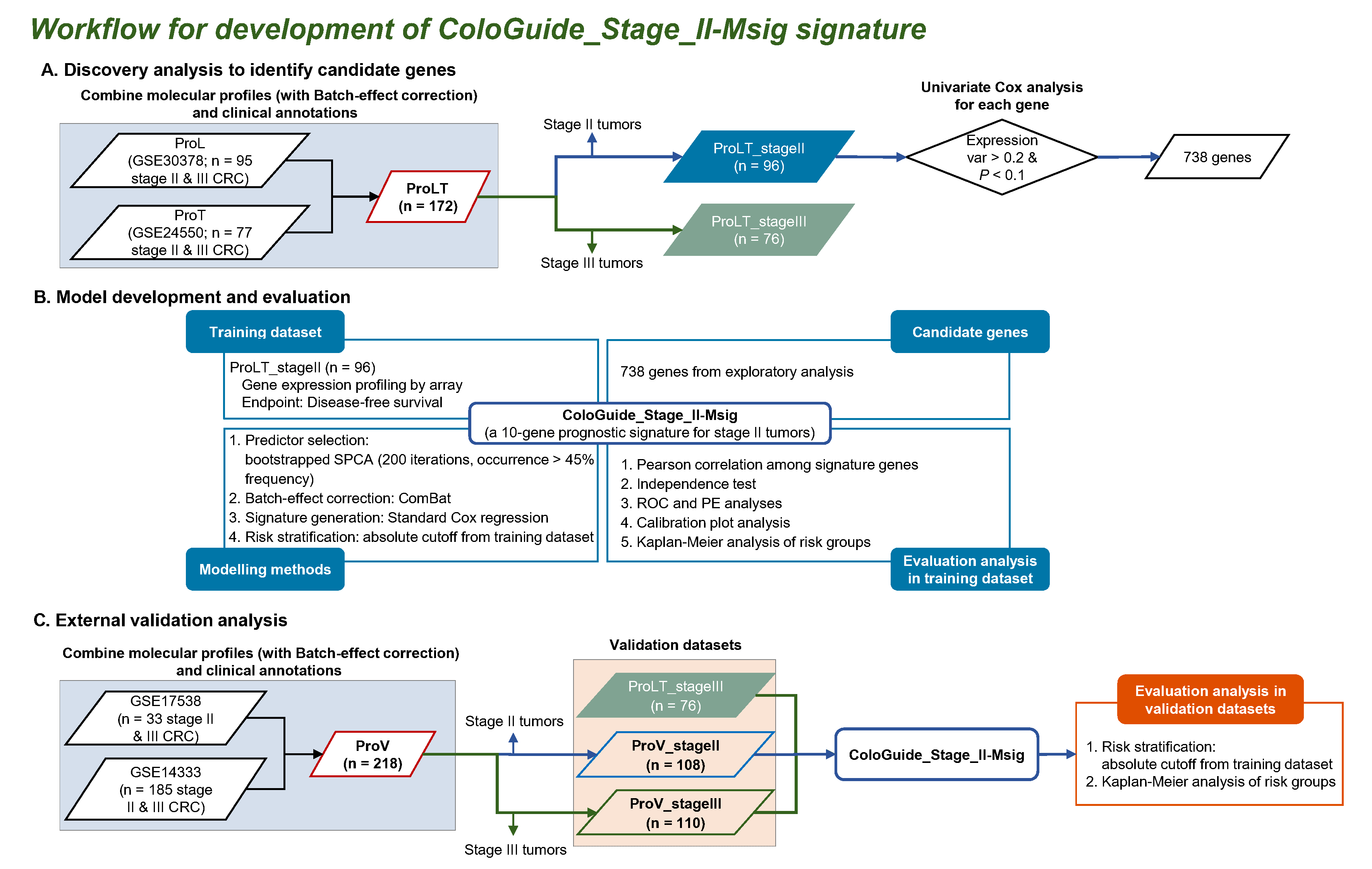
Example 1: ColoGuide_Stage_II-Msig
Example 2: ColoGuide_Stage_III-Msig
Candidate predictors
Predictor selection by a bootstrapping approach
Predictor selection methods
Bootstrapping parameters
Bootstrap iterations
Frequency of selected predictors (%)
Options
Batch effect correction
Signature generation
Modelling data
Time intervals (months)
Risk stratification based on signature score
Cutoff to subgroup patients
Subgroup variable for Kaplan-Meier analysis
Subgroup variable
Clinicopathological variables
Variable combination
Variable selection
Independence test
Interaction test
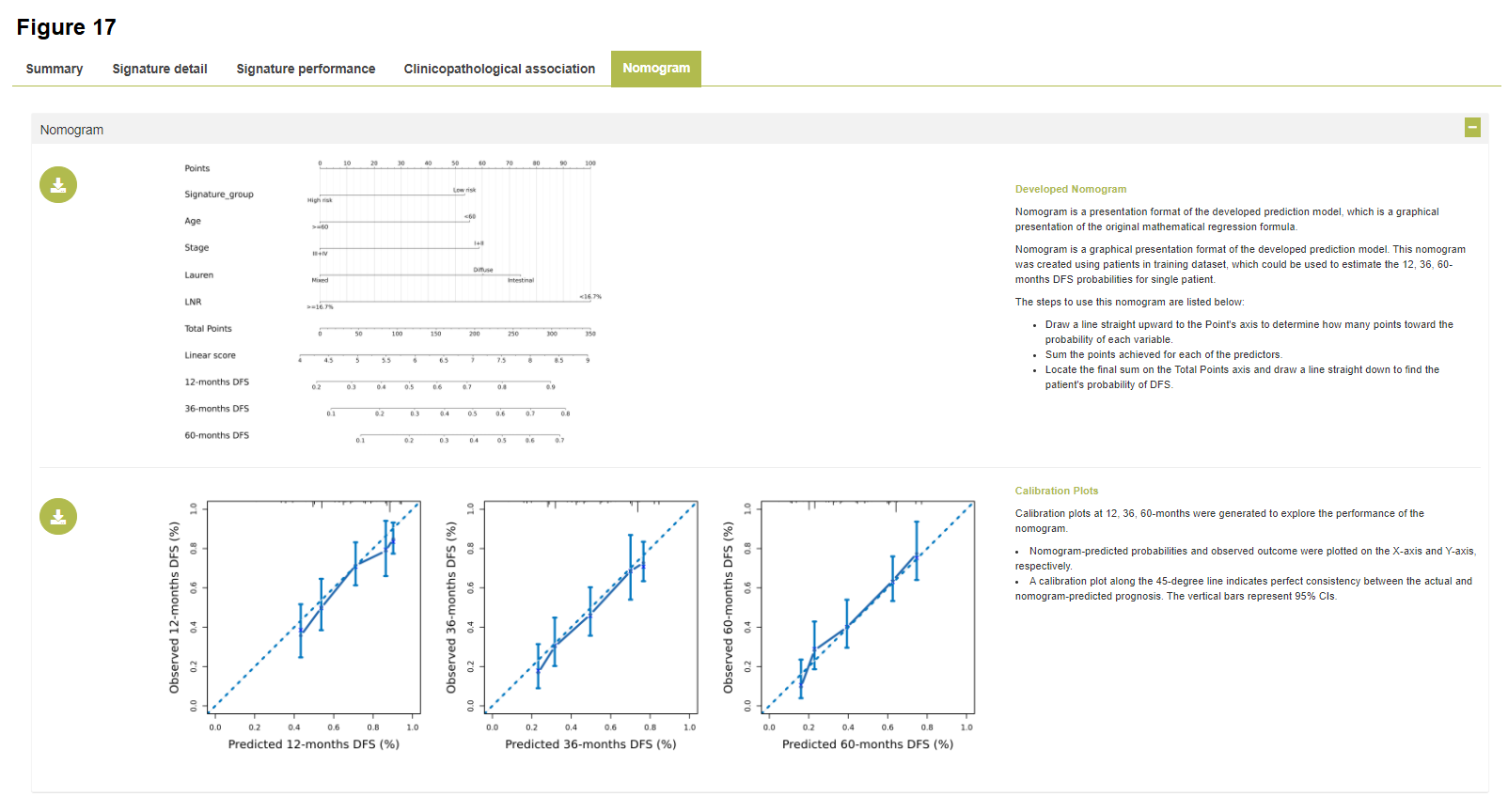
Nomogram
Clinicopathological association analysis
Results
The results of signature development, validation and evaluation are categorized in multiple tabs as below:
Copyright © 2023
Center for Cancer Bioinformatics
All Rights Reserved.
Peking University Cancer Hospital & Institute, Peking University Health Science Center

Step 1
This module can predicts prognosis or treatment response for a new case, by calculating the risk score (RS) and event probabilities at specified intervals using a selected prognostic/predictive biomarker.
Please note:
- For molecular signatures, users need to upload molecular profiles with all molecular variables utilized in the selected signature.
- For traditional signatures, users need to provide value of each clinicopathological predictor utilized in the model (i.e. Nomogram).
- A unique patient ID is required.
Step 2
Datasets suitable for the selected signature are listed in the table below for selection, which will be used as reference cohort(s) for indicating ranks of predicted risk score (RS) of this case.
Results
In the case of a prognostic signature, risk group and specified event probabilities (e.g. 1, 3, and 5-year) of the studied case will be predicted with visualization output.
In the case of a predictive signature, it will predict whether a patient might benefit from treatment of interest, based on its assigned risk group using the RS cutoff defined by the training dataset.
An risk score (RS) ranking feature is provided to indicate the percentile rank of the predicted RS for this individual, in the RS distribution of each selected dataset (with the same clinical endpoint and all utilized predictors).
Please note:
- Registered users can save their histories in the platform for future access.
Copyright © 2023
Center for Cancer Bioinformatics
All Rights Reserved.
Peking University Cancer Hospital & Institute, Peking University Health Science Center

Copyright © 2023
Center for Cancer Bioinformatics
All Rights Reserved.
Peking University Cancer Hospital & Institute, Peking University Health Science Center

Forgotten Password
Please enter the email address that you used for registration of your ReProMSig account and submit. We will send a email to that address containing a initial password that you can use to reset your password within 24 hours.
Forgotten Password
Forgotten Password
Login
Update Password
Copyright © 2023
Center for Cancer Bioinformatics
All Rights Reserved.
Peking University Cancer Hospital & Institute, Peking University Health Science Center

Disclaimer
ReProMSig is an online platform for developing and validating a Reproducible Prognosis Molecular Signature in a transparent and reproducible way. This website is free and open to academic users and there is no login requirement. By using this website, you agree to the entire content of this statement.
Copyright © 2023
Center for Cancer Bioinformatics
All Rights Reserved.
Peking University Cancer Hospital & Institute, Peking University Health Science Center