Introduction
Protein Interaction Network Analysis (PINA) platform is an
integrated platform for protein interaction network construction, filtering, analysis,
visualization, and management.
It integrates protein-protein interaction (PPI) data from public curated databases
and builds a complete, non-redundant protein interaction dataset for six model organisms.
In particular, it provides a variety of built-in tools to filter and analyze the networks
for gaining biological and functional insights into the network.
PINA v2.0 Mining interactome modules.
PINA v3.0 Revealing tumour type-specific insights from PPI networks.
News:
26 Sept. 2020 : The normalization strategy of mRNA expression data was updated to be comparable with other published studies,
and the tutorial step-by-step case study was updated accordingly. The Data & Statistics page was redesigned to be more informative.
Usage
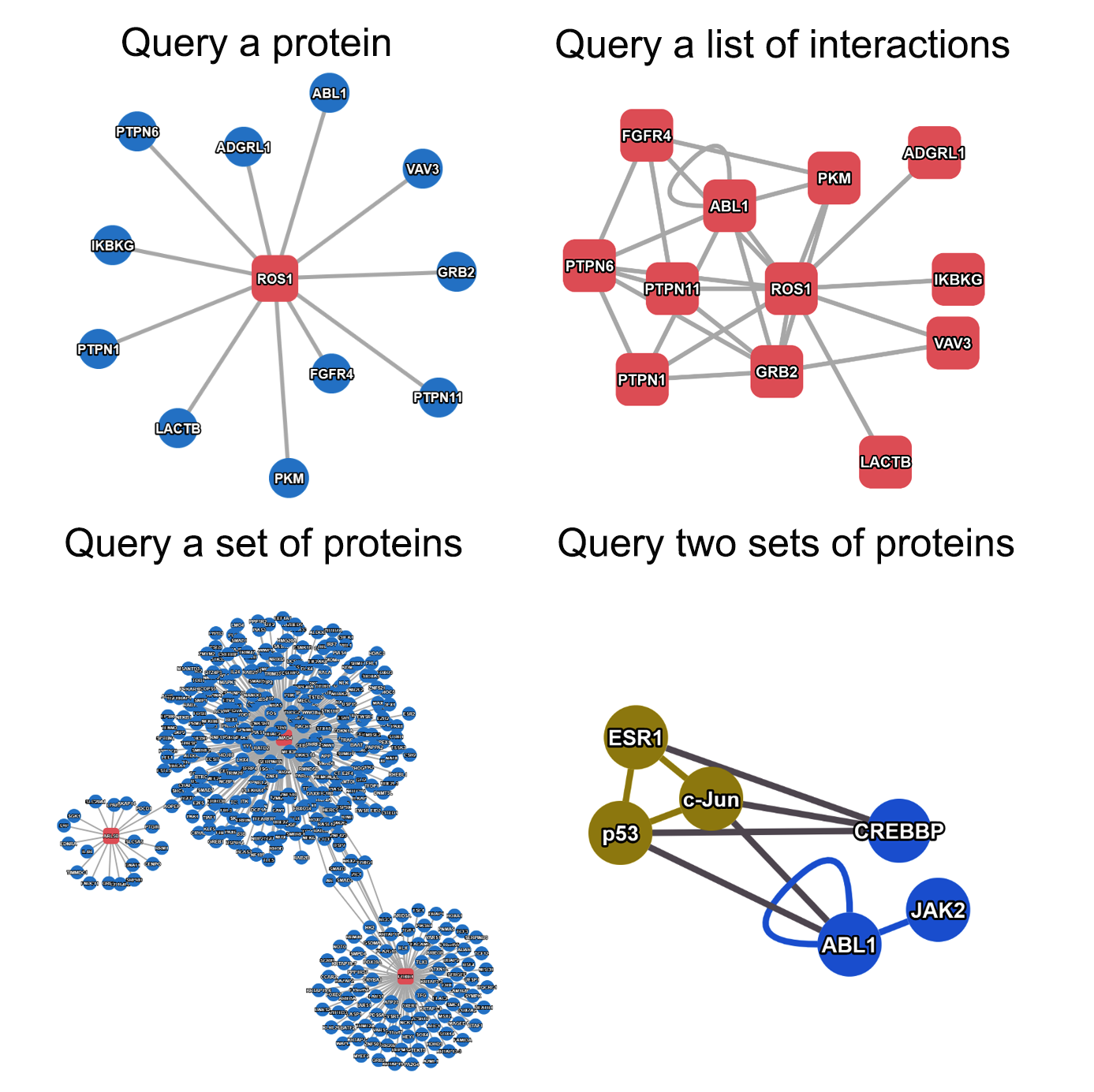
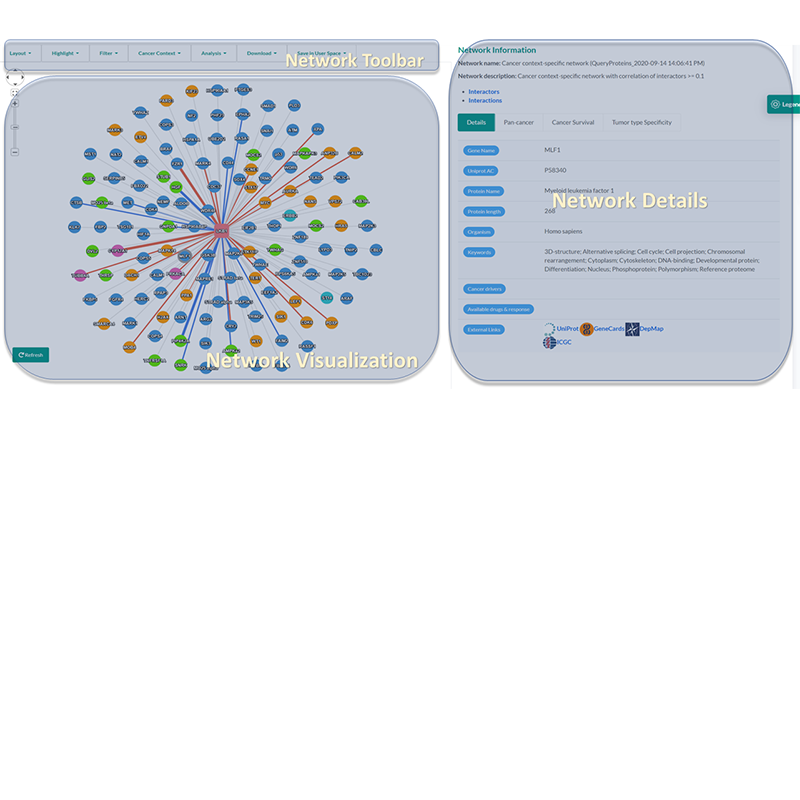
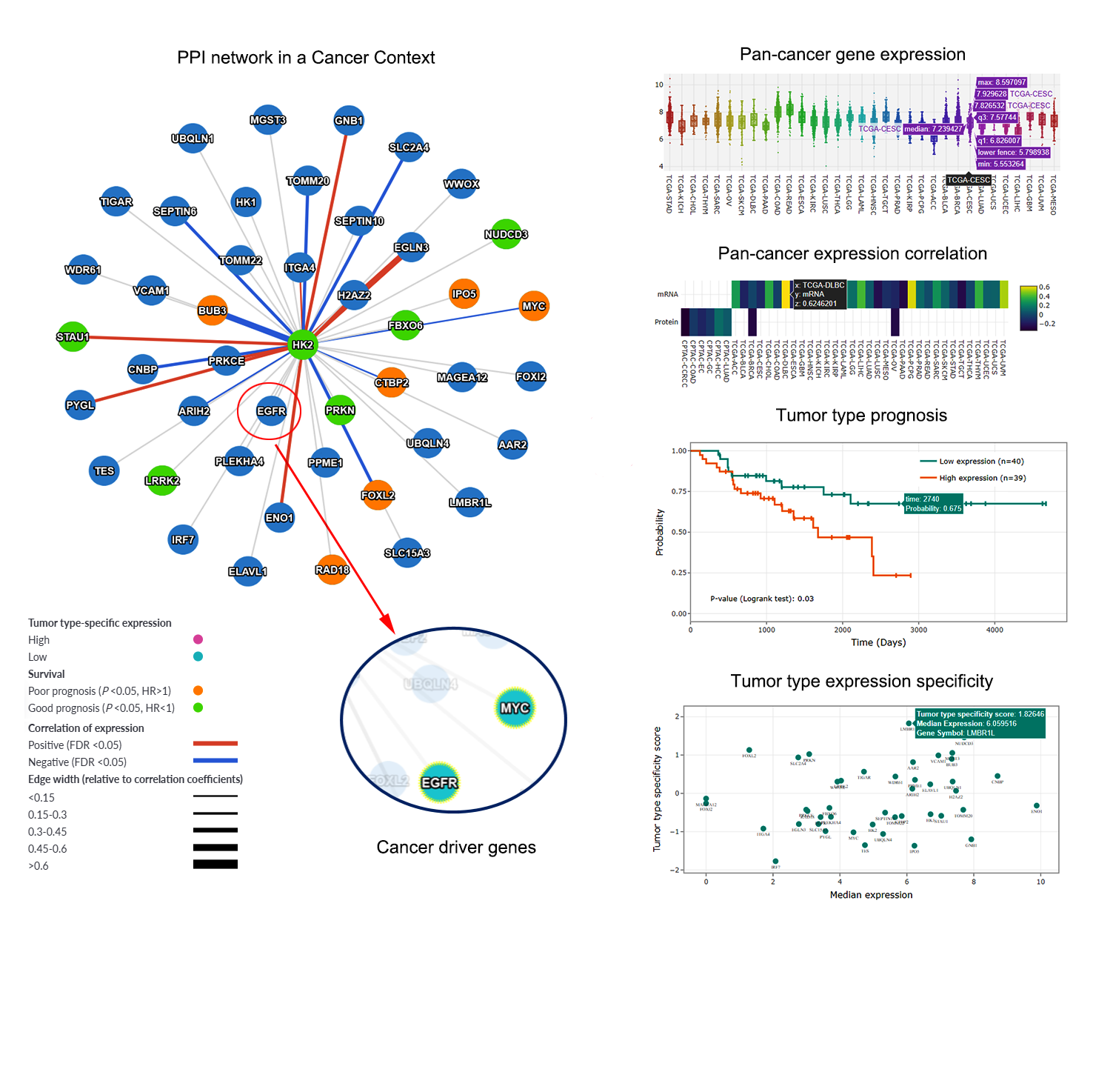
- to infer network proteins with tumor type expression specificity.
- to identify candidate prognosis biomarkers, putative mutational cancer drivers, and therapeutic targets in a network, for a specific cancer type.
- to identify pairs of co-expressing interacting proteins across cancer types.
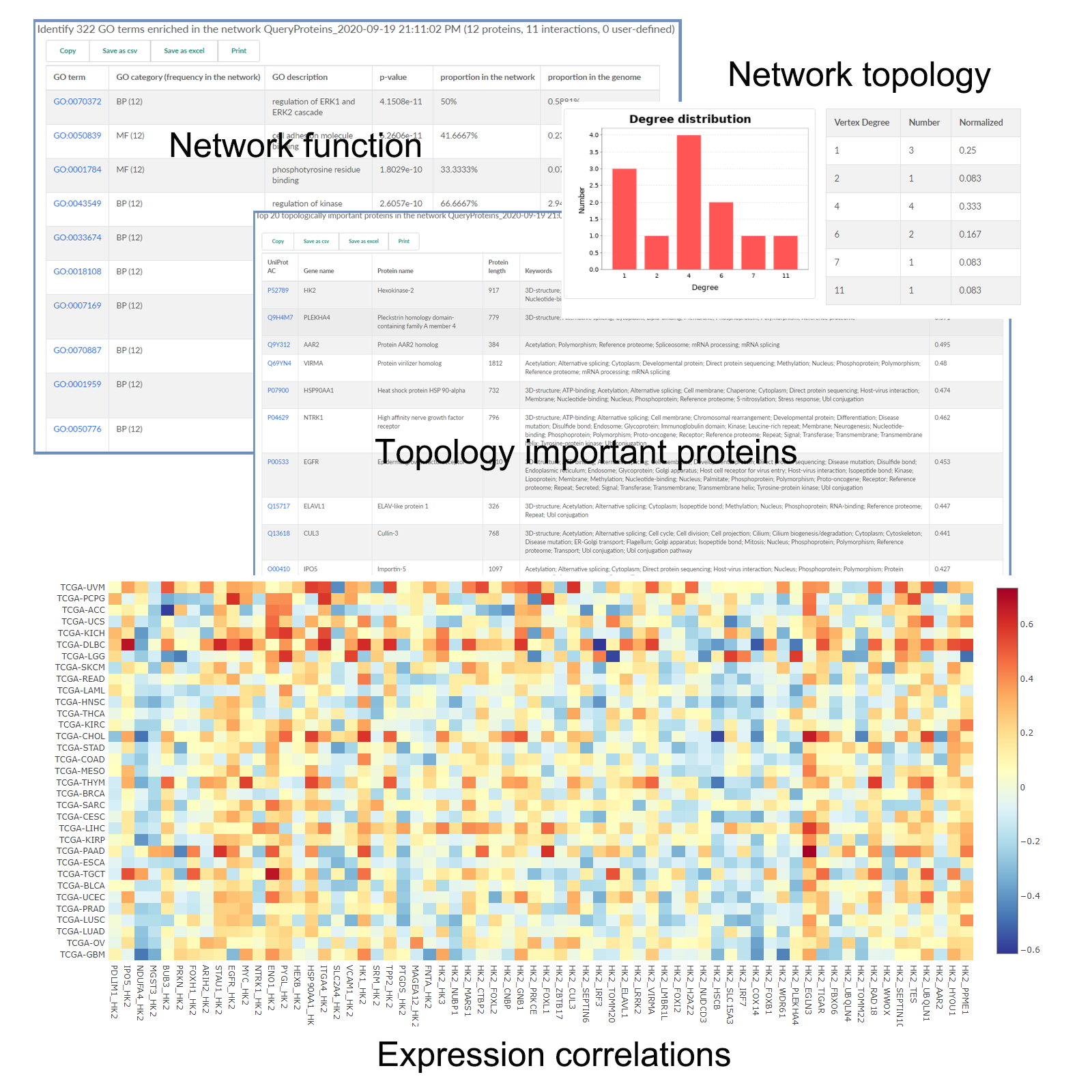
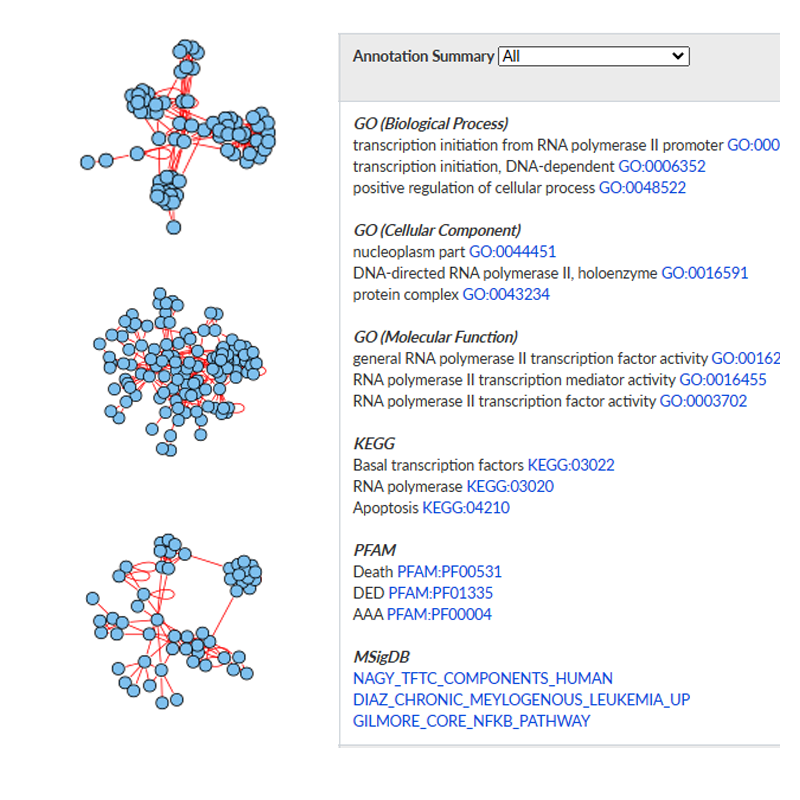
- The identified modules have been comprehensively annotated with known knowledge from Gene Ontology, KEGG pathways, PFAM domains, and MSigDB.
- Users can either simply search modules or identify enriched modules from the list of query proteins.
Check the online Tutorial for a more detailed explanation of theuser interface and results.
Citing PINA
Du, Y., Cai, M., Xing, X., Ji, J., Yang, E. and Wu, J. (2021)
PINA 3.0: mining cancer interactome. Nucleic Acids Res, 49, D1351-D1357.
[Full
text]
Cowley, M.J., Pinese, M., Kassahn, K.S., Waddell, N., Pearson, J.V., Grimmond, S.M.,
Biankin, A.V., Hautaniemi, S. and Wu, J. (2012)
PINA v2.0: mining interactome modules. Nucleic Acids Res, 40,
D862-865.
[Full
text]
Wu, J., Vallenius, T., Ovaska, K., Westermarck, J., Makela, T.P. and Hautaniemi, S.
(2009)
Integrated network analysis platform for protein-protein interactions, Nature
methods, 6, 75-77.
[Abstract]
